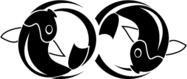Personalization
Anatomical Modeling
Current tomographic imaging modalities allow the geometrically accurate reconstruction of an individual's cardiac anatomy. A key objective is the conception of methods which allow to turn such tomographic image stacks into discrete anatomical representations which can be used for modeling cardiac physiology.
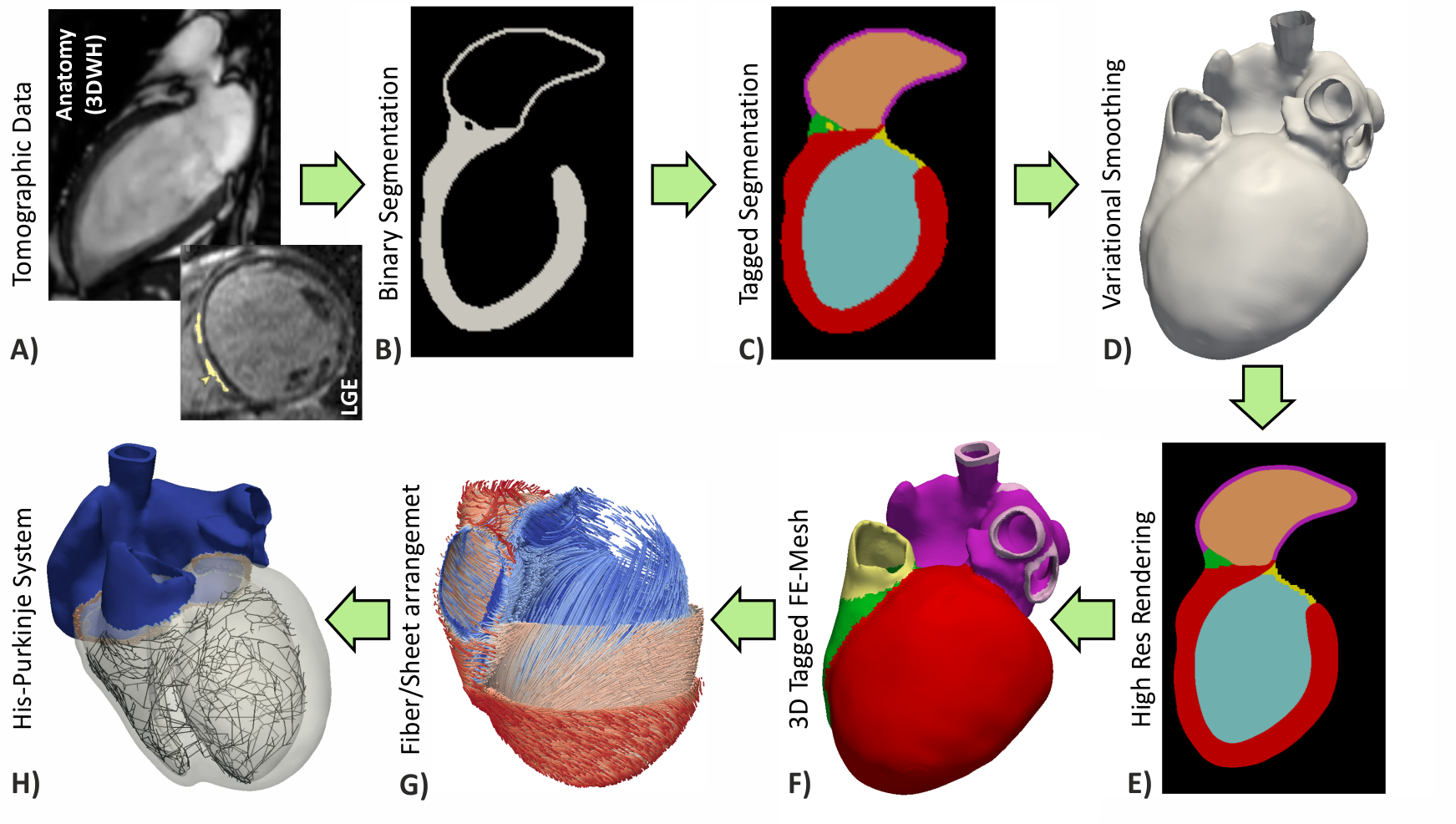
Figure: Workflow for the generation of cardiac models from medical image data
Image Based Mesh Generation
The conversion of tomographic imaging data into discrete finite element models relies upon model generation pipelines as illustrated in the figure above. The desire to translate the image data into finite element space and tailor it to specific physics may impose strikingly different requirements. Electrophysiology models feature fast transients in time and steep wave fronts in space, which makes high spatio-temporal resolutions necessary to capture these dynamics. In contrast, due to the smoother spatio-temporal characteristics of mechanical deformation, numerical constraints upon discretization are less severe. Preferably, relevant anatomical details and a sufficiently smooth surface of organs need to be preserved. Moreover, anatomical classification of voxel groups (e.g. torso, lung, heart, ...), obtained during the segmentation process, need to be transferred. These requirements can be met using an image-based unstructured mesh generation technique which produces conformal, boundary-fitted, hexahedra-dominant and adaptive meshes in a fully automatic fashion.
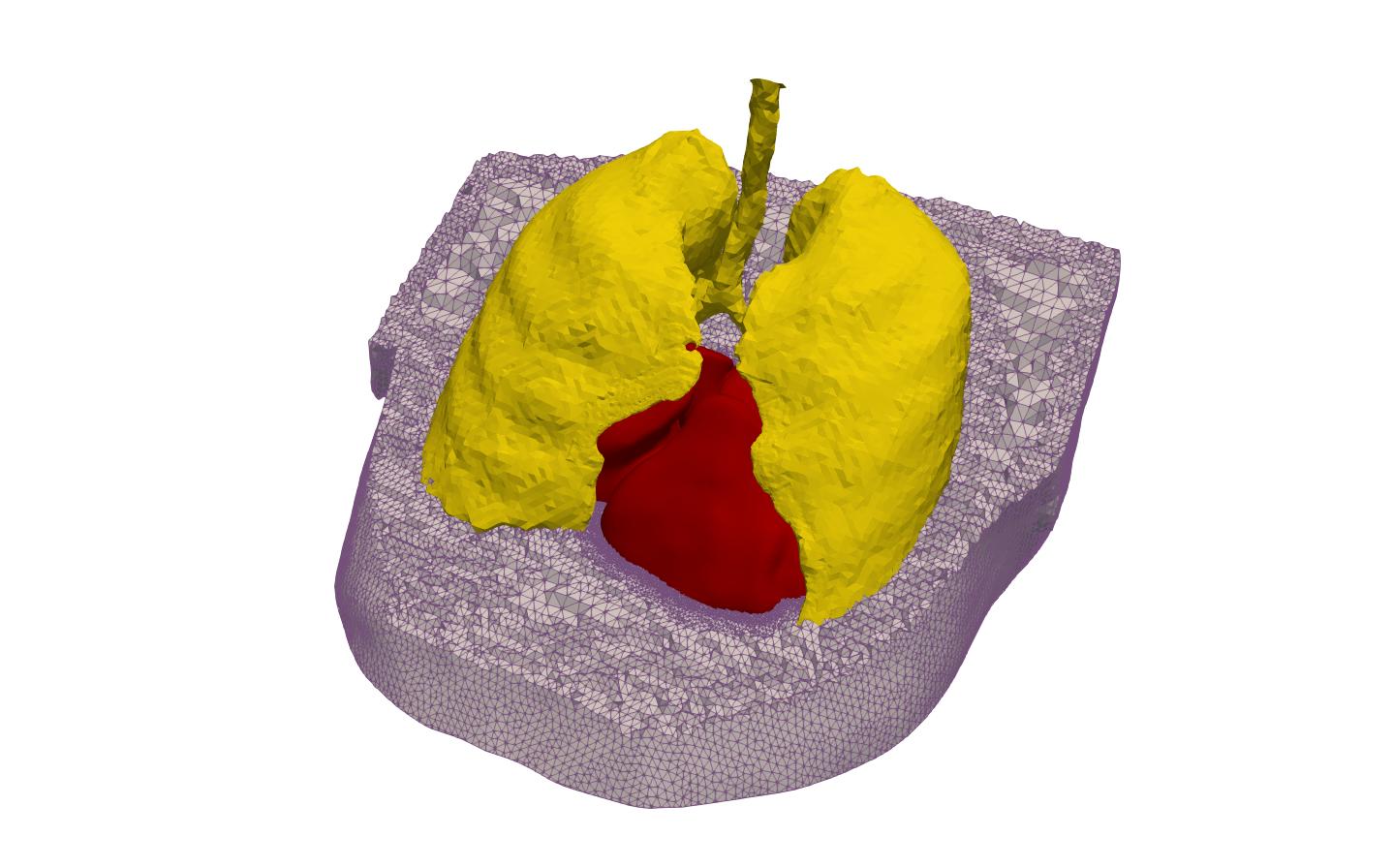
Figure: Torso model
Rule-Based Fiber Algorithm
Geometric models derived from imaging data do not include information on the distribution of fiber orientations. Knowledge of the structural anisotropy, however, is essential for modelling the electrical conduction and active force generation. In absence of histology or ex-vivo diffusion tensor MRI, mathematical rules are applied to grade up personalized anatomical models.
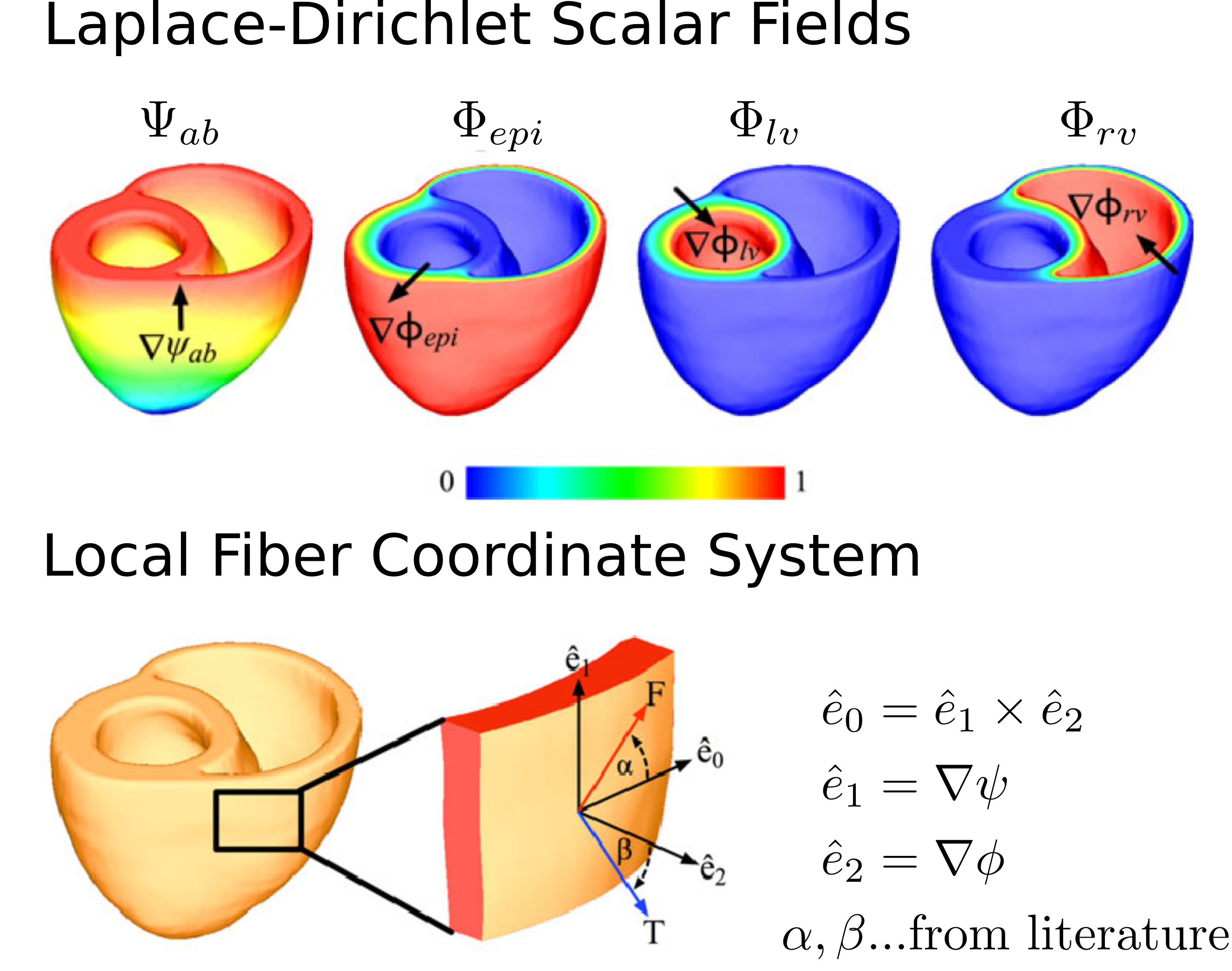
Figure: Laplace-Dirichlet Scalar Fields
Universal Ventricular Coordinates (UVC)
Performing studies with anatomically accurate models of patients has become the state-of-the-art in computational modeling in cardiac electro-mechanics. Their application in a clinical context, enriched with patient-specific functionalization for diagnostics and therapy planning, shows high promise. Tuning spacial and temporal parameters involves labor and time-intensive workflows with the need of efficient and easily applicable data structures. For this sake UVC coordinates were introduced to quickly navigate within a ventricular geometry or map data between comparable geometries.
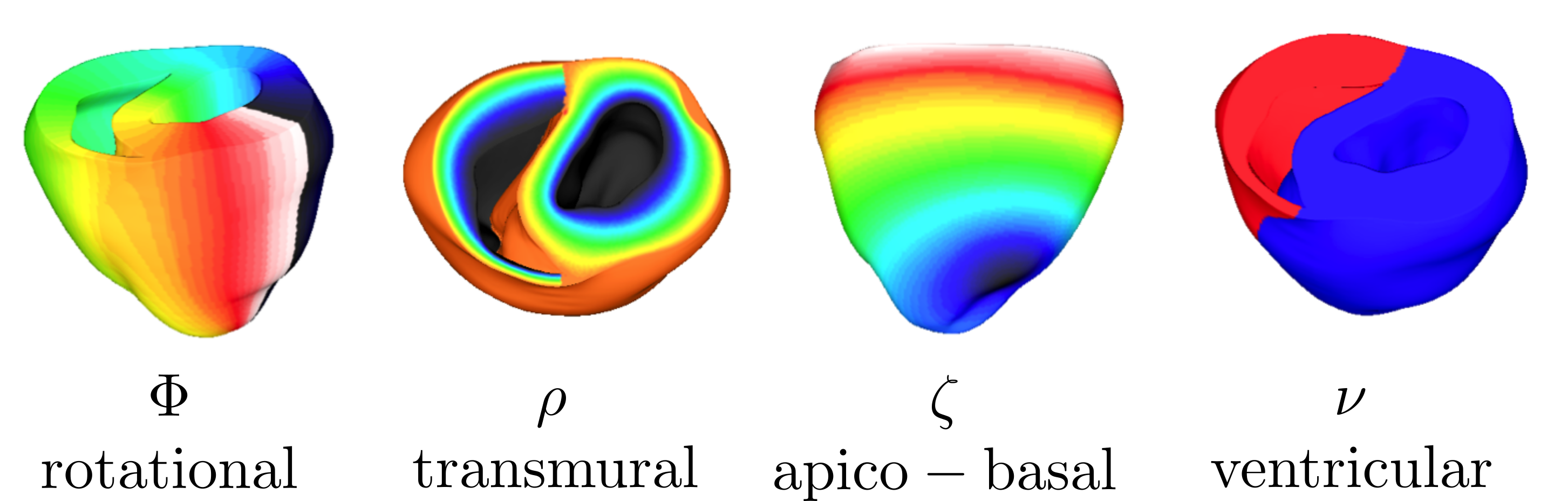
Figure: UVC coordinates
Digital Twinning
A digital twin is a digital replica of an individual person.
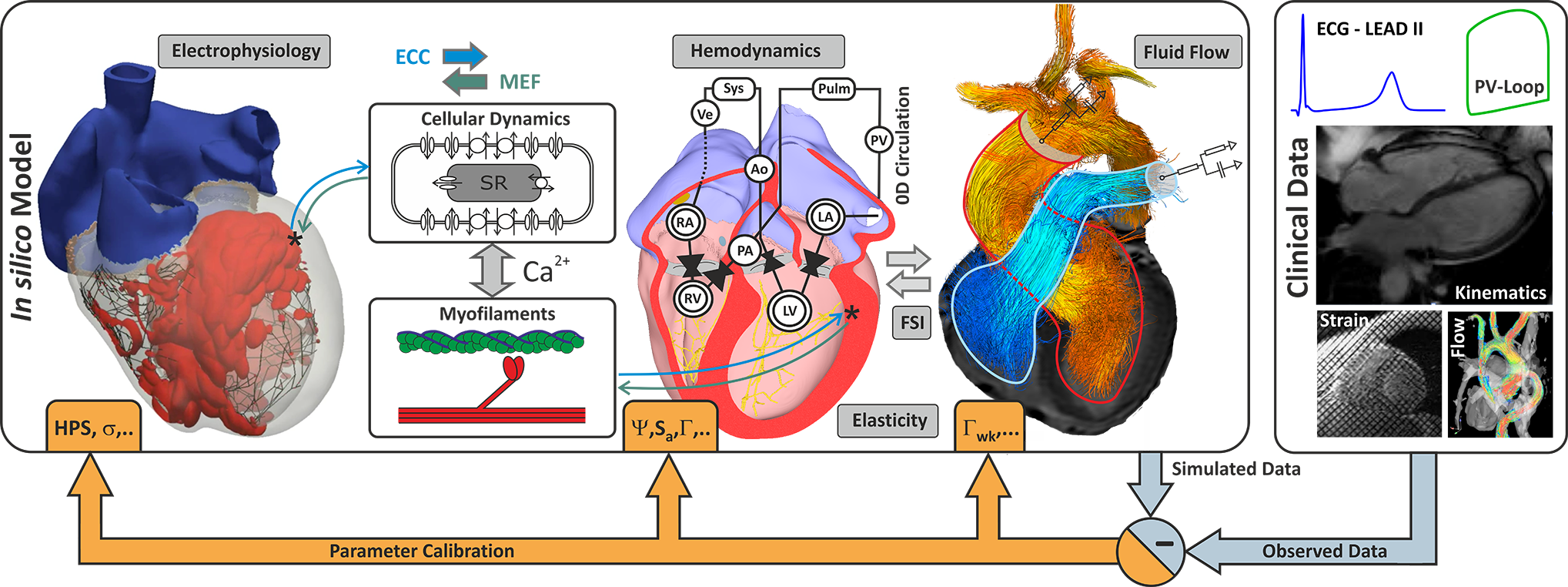
Figure: Personalization of model parameters.
Video 1: Extracellular potentials in heart and torso
Mechanical Personalization
The detailed workflow used for parameterization of electro-mechanical LV models is described in [Augustin et al.,2016] , personalizing passive and active mechanical model components as well as circulatory components. Constitutive relations accounting for the passive mechanical properties of the cardiac tissue are represented in terms of the Guccione Model . From the anatomical models at end-diastolic state, a stress-free reference configuration is computed using default material parameters and end-diastolic pressure in the LV. This is accomplished by unloading the geometry using a backward displacement method. The EDPVR is fitted to the empiric Klotz EDPVR using default values from literature for the parameters [Guccione et al.,1991] adapting only the stiffness parameter \(C_{\mathrm{Guc}}\). A simplified phenomenological contractile model was used to represent active stress generation , which was fitted using default values of \(\tau_\mathrm{C}\), \(\hat{S}_{\rm a}\), \(\tau_\mathrm{R}\) as initial guess. \(T_\mathrm{dur}\) was initialised with the RT interval observed in the ECG. A linear mapping was used to correct the active stress model parameters. When measured pressure traces \(p_\mathrm{lv,m}(t)\) were not available, only \(\hat{S}_{\rm a}\) was iteratively adjusted by a fixed-point iteration \(\hat{S}_{{\rm a},i+1} = \hat{S}_{{\rm a},i} \cdot \hat{p}_{\mathrm{lv}}/\hat{p}_{\mathrm{lv},i}\). Otherwise, rate of rise \(\tau_\mathrm{C}\) and decline \(\tau_\mathrm{R}\) of active stress were adjusted accordingly using \(\frac{p_{\rm lv,m}}{dt}|_\mathrm{max}\) and \(\frac{p_{\rm lv,m}}{dt}|_\mathrm{min}\) as references and fitted in an similar manner. Circulatory components in terms of the three-element Windkessel model were identified separately using a global-local optimization approach. Both workflows are visualized below.